3 mins of Machine Learning: Ada-boost
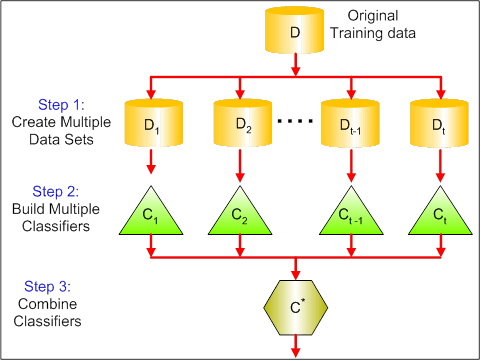
1 Introduction
As an ensambled method, boosting refers to a general and provably effective method of producing a very accurate prediction rule by combining rough and moderately inaccurate simple predication models. It is an iterative procedure to adaptively change distribution of training data by focusing more on previously misclassified records. Initially, all N records are assigned equal weights. And unlike bagging, weights may change at the end of each boosting round. The AdaBoost algorithm, introduced in 1995 by Freund and Schapire, solved many of the practical difficulties of the earlier boosting algorithms. In this post, I will write my own Adaboost and compare its performance with other models.
2 Adaboost Algorithm
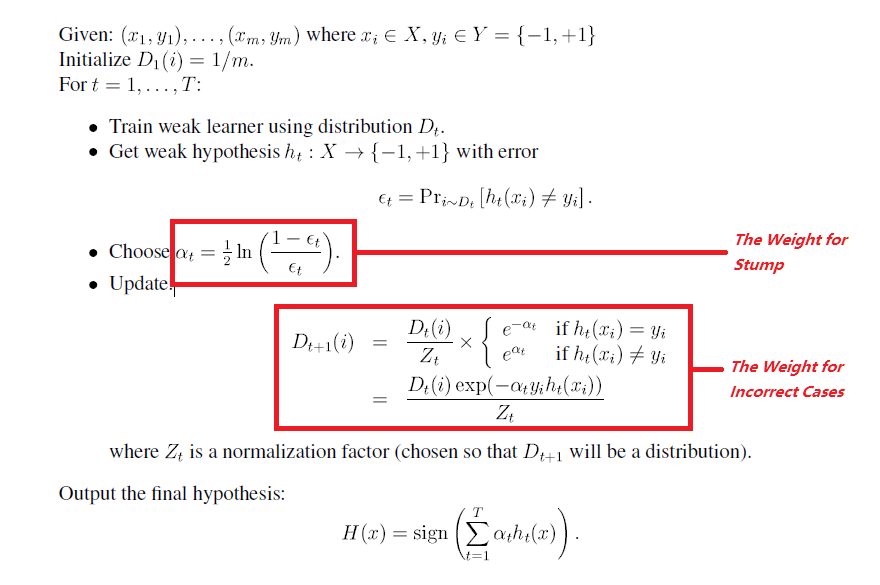
The main idea of Adaboost is to maintain a distribution or set of weights over the training set. Initially, all weights are set equally, but on each round, the weights of incorrectly classified examples are increased so that the weak learner is forced to focus on the hard examples in the training set. For more details you can check out this paper
Here is the algorithm
3 R Code
In my code, I use regression tree with depth = 1 to be my stump
library(rpart)
## train_feature: train data's feature
## response: train data's response, it needs to be binary for my code
## test_feature: test data's feature
My_Adaboost <- function(train_feature, response, test_feature, t) {
n = nrow(train_feature)
m = nrow(test_feature)
w = rep(1 / n, n)
alpha = rep(0, t)
result = rep(0, m)
stump <- vector("list", t)
for (i in 1:t) {
ctrl <- rpart.control(cp = 0,
maxdepth = 1,
xval = 0)
stump[[i]] = rpart(
factor(response) ~ .,
data = train_feature,
method = "class",
weights = w,
control = ctrl
)
result_on_train = as.numeric(as.character(as.vector(
predict(stump[[i]], train_feature, type = "class")
)))
epsilon = w %*% (1 * (result_on_train != response)) / sum(w)
alpha[i] = 0.5 * (log((1 - epsilon) / epsilon))
sign = ifelse(result_on_train == response, -1, 1)
exp_part = exp(sign * alpha[i])
z = w %*% exp_part
w = (w * exp_part) / rep(z, n)
}
for (j in 1:t) {
f = as.numeric(as.character(as.vector(
predict(stump[[j]], test_feature, type = "class")
)))
new = alpha[j] * f
result = new + result
}
pred = sign(result)
return(pred)
}4 Performance
## Change response
attach(stagec)
stagec= na.omit(stagec)
stagec1=stagec[,-c(1)]
stagec1$pgstat=ifelse(stagec$pgstat==0,-1,1)
head(stagec1)## pgstat age eet g2 grade gleason ploidy
## 1 -1 64 2 10.26 2 4 diploid
## 3 1 59 2 9.99 3 7 diploid
## 4 1 62 2 3.57 2 4 diploid
## 5 1 64 2 22.56 4 8 tetraploid
## 6 -1 69 1 6.14 3 7 diploid
## 9 1 73 2 11.77 3 6 diploid##Train test data spilting
train_index=sample(c(1:nrow(stagec1)), 120)
train=stagec1[train_index,]
test=stagec1[-train_index,]
test_feature=test[,-c(1)]
train_feature=train[,-c(1)]
test_response=test[,1]
train_response=train[,1]## Adaboost
pred=My_Adaboost(train_feature = train_feature,response = train_response,test_feature = test_feature,t=100)
mean(1*(test_response==pred))## [1] 0.6428571Now, let’s compare the test error to the test
error from a single classification tree that is learned using rpart()
## Comparing with a single classification tree
model=rpart(factor(train_response) ~.,
data = train_feature, method = "class")
pred_original=predict(model,test_feature,type="class")
mean(1*(test_response==pred_original))## [1] 0.5714286